In this guide, you'll learn how to set up distributed hyperparameter tuning for machine learning models across multiple nodes using Kubernetes. You'll use Optuna, a bayesian optimization library, to fine-tune models built with popular libraries like scikit-learn, XGBoost, PyTorch, and TensorFlow/Keras.
To orchestrate all the trials, you'll use Neon Postgres, a serverless postgres database. The combination of Neon Postgres, Kubernetes, and Docker allows for scalable, distributed hyperparameter tuning, simplifying the orchestration and management of complex machine learning workflows.
Prerequisites
Before you begin, ensure you have the following tools and services set up:
Neon Serverless Postgres: To provision and manage your serverless PostgreSQL database. If you don't have an account yet, sign up here.Minikube: For running a local Kubernetes cluster. You can install it by following the official Minikube installation guide.kubectl: Kubernetes command-line tool for interacting with your cluster. Follow the kubectl installation instructions to get started.Docker: For containerizing your applications. If you don't have it installed, check out the Docker installation guide.Python: To create, train, and optimize machine learning models. You can download Python from the official website.
Overview
Hyperparameters are essential to machine learning model performance. Unlike parameters learned during training, hyperparameters—like learning rates, batch sizes, or the number of layers in a neural network—need to be set in advance. Tuning these hyperparameters effectively can greatly improve model performance, squeezing out the last bit of accuracy or reducing training time.
Bayesian optimization offers an efficient method for hyperparameter tuning. Unlike traditional approaches like grid or random search, Bayesian optimization builds a probabilistic model of the objective function to help it sample which hyperparameters to test next, which reduces the number of experiments needed, saving time and compute.
Distributing hyperparameter tuning across multiple nodes allows each trial to run independently on its own machine, enabling multiple configurations to be tested simultaneously and speeding up the search process. However, these nodes need to be coordinated, and a serverless database like Neon Postgres is perfect. Neon offers a pay-as-you-go model that minimizes costs during idle periods,but scales when the workload demands it. This database will maintain the state of the hyperparameter tuning process, storing the results of each trial and coordinating the distribution of new trials to available nodes.
These nodes are managed using Kubernetes, a container orchestration platform, which enables the same task to run concurrently across multiple nodes, allowing each node to handle a separate trial independently. It can also manage resources per node, and reboot nodes that fail, allowing for a fault tolerant training process.
In this guide, you will combine Optuna, Kubernetes, and Neon Postgres to create a scalable and cost-effective system for distributed tuning of your PyTorch, TensorFlow/Keras, scikit-learn, and XGBoost models.
Hyperparameter Tuning
Optuna organizes hyperparameter tuning into studies, which are collections of trials. A study represents a single optimization run, and each trial within the study corresponds to a set of hyperparameters to be evaluated. Optuna uses a study to manage the optimization process, keeping track of the trials, their results, and the best hyperparameters found so far.
When you create a study, you can specify various parameters like the study name, the direction of optimization (minimize or maximize), and the storage backend. The storage backend is where Optuna stores the study data, including the trials and their results. By using a persistent storage backend like Neon Postgres, you can save the state of the optimization process, allowing all nodes to access the same study and coordinate the tuning process.
A study is created like so :
if __name__ == "__main__":
study = optuna.create_study(
study_name="sklearn_example",
storage=os.environ["DATABASE_URL"],
load_if_exists=True,
direction="maximize",
)
study.optimize(objective, n_trials=100)In the case of the distributed training, the load_if_exists parameter is set to True to load an existing study if it already exists, allowing nodes to join the optimization process.
Based on your machine learning library of choice, you can define an objective function that takes a trial object as input and returns a metric to optimize. Let's dive into each library and see how to define the objective function for scikit-learn, XGBoost, PyTorch, and TensorFlow/Keras models using test datasets.
To follow along, name your python script hyperparam_optimization.py.
sklearn
ScikitLearn has a very wide range of models in its library, but in this you will be comparing a Support Vector Classifier and a Random Forest Classifier, and their hyperparameter configurations. For the SVC, you will optimize the strength of the regularization parameter C, while for the RF, you will optimize the maximum depth of the trees max_depth.
import os
import optuna
import sklearn.datasets
import sklearn.ensemble
import sklearn.model_selection
import sklearn.svm
def objective(trial):
iris = sklearn.datasets.load_iris()
x, y = iris.data, iris.target
classifier_name = trial.suggest_categorical("classifier", ["SVC", "RandomForest"])
if classifier_name == "SVC":
svc_c = trial.suggest_float("svc_c", 1e-10, 1e10, log=True)
classifier_obj = sklearn.svm.SVC(C=svc_c, gamma="auto")
else:
rf_max_depth = trial.suggest_int("rf_max_depth", 2, 32, log=True)
classifier_obj = sklearn.ensemble.RandomForestClassifier(
max_depth=rf_max_depth, n_estimators=10
)
score = sklearn.model_selection.cross_val_score(classifier_obj, x, y, n_jobs=-1, cv=3)
accuracy = score.mean()
return accuracy
if __name__ == "__main__":
study = optuna.create_study(
study_name="sklearn_example",
storage=os.environ["DATABASE_URL"],
load_if_exists=True,
direction="maximize",
)
study.optimize(objective, n_trials=100)
print(study.best_trial)xgboost
Gradient Boosting is king in the world of tabular data, and XGBoost is one of the most popular libraries for this task. However, these models are especially sensitive to hyperparameter choice. In this example, you will optimize the booster type, regularization weights, sampling ratios, and tree complexity parameters.
import numpy as np
import os
import optuna
import sklearn.datasets
import sklearn.metrics
from sklearn.model_selection import train_test_split
import xgboost as xgb
def objective(trial):
(data, target) = sklearn.datasets.load_breast_cancer(return_X_y=True)
train_x, valid_x, train_y, valid_y = train_test_split(data, target, test_size=0.25)
dtrain = xgb.DMatrix(train_x, label=train_y)
dvalid = xgb.DMatrix(valid_x, label=valid_y)
param = {
"verbosity": 0,
"objective": "binary:logistic",
# use exact for small dataset.
"tree_method": "exact",
# defines booster, gblinear for linear functions.
"booster": trial.suggest_categorical("booster", ["gbtree", "gblinear", "dart"]),
# L2 regularization weight.
"lambda": trial.suggest_float("lambda", 1e-8, 1.0, log=True),
# L1 regularization weight.
"alpha": trial.suggest_float("alpha", 1e-8, 1.0, log=True),
# sampling ratio for training data.
"subsample": trial.suggest_float("subsample", 0.2, 1.0),
# sampling according to each tree.
"colsample_bytree": trial.suggest_float("colsample_bytree", 0.2, 1.0),
}
if param["booster"] in ["gbtree", "dart"]:
# maximum depth of the tree, signifies complexity of the tree.
param["max_depth"] = trial.suggest_int("max_depth", 3, 9, step=2)
# minimum child weight, larger the term more conservative the tree.
param["min_child_weight"] = trial.suggest_int("min_child_weight", 2, 10)
param["eta"] = trial.suggest_float("eta", 1e-8, 1.0, log=True)
# defines how selective algorithm is.
param["gamma"] = trial.suggest_float("gamma", 1e-8, 1.0, log=True)
param["grow_policy"] = trial.suggest_categorical("grow_policy", ["depthwise", "lossguide"])
if param["booster"] == "dart":
param["sample_type"] = trial.suggest_categorical("sample_type", ["uniform", "weighted"])
param["normalize_type"] = trial.suggest_categorical("normalize_type", ["tree", "forest"])
param["rate_drop"] = trial.suggest_float("rate_drop", 1e-8, 1.0, log=True)
param["skip_drop"] = trial.suggest_float("skip_drop", 1e-8, 1.0, log=True)
bst = xgb.train(param, dtrain)
preds = bst.predict(dvalid)
pred_labels = np.rint(preds)
accuracy = sklearn.metrics.accuracy_score(valid_y, pred_labels)
return accuracy
if __name__ == "__main__":
study = optuna.create_study(
study_name="xgboost_example",
storage=os.environ["DATABASE_URL"],
load_if_exists=True,
direction="maximize",
)
study.optimize(objective, n_trials=100)PyTorch
PyTorch is now the defacto library for deep learning research, and its flexibility makes it a popular choice for many machine learning tasks. In this example, you will optimize the number of layers, hidden units, and dropout ratios in a feedforward neural network for the FashionMNIST dataset, a popular benchmark for image classification.
import os
import optuna
from optuna.trial import TrialState
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torch.utils.data
from torchvision import datasets
from torchvision import transforms
DEVICE = torch.device("cpu")
BATCHSIZE = 128
CLASSES = 10
DIR = os.getcwd()
EPOCHS = 10
N_TRAIN_EXAMPLES = BATCHSIZE * 30
N_VALID_EXAMPLES = BATCHSIZE * 10
def define_model(trial):
n_layers = trial.suggest_int("n_layers", 1, 3)
layers = []
in_features = 28 * 28
for i in range(n_layers):
out_features = trial.suggest_int("n_units_l{}".format(i), 4, 128)
layers.append(nn.Linear(in_features, out_features))
layers.append(nn.ReLU())
p = trial.suggest_float("dropout_l{}".format(i), 0.2, 0.5)
layers.append(nn.Dropout(p))
in_features = out_features
layers.append(nn.Linear(in_features, CLASSES))
layers.append(nn.LogSoftmax(dim=1))
return nn.Sequential(*layers)
def get_mnist():
train_loader = torch.utils.data.DataLoader(
datasets.FashionMNIST(DIR, train=True, download=True, transform=transforms.ToTensor()),
batch_size=BATCHSIZE,
shuffle=True,
)
valid_loader = torch.utils.data.DataLoader(
datasets.FashionMNIST(DIR, train=False, transform=transforms.ToTensor()),
batch_size=BATCHSIZE,
shuffle=True,
)
return train_loader, valid_loader
def objective(trial):
model = define_model(trial).to(DEVICE)
# Generate the optimizers.
optimizer_name = trial.suggest_categorical("optimizer", ["Adam", "RMSprop", "SGD"])
lr = trial.suggest_float("lr", 1e-5, 1e-1, log=True)
optimizer = getattr(optim, optimizer_name)(model.parameters(), lr=lr)
train_loader, valid_loader = get_mnist()
for epoch in range(EPOCHS):
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.view(data.size(0), -1).to(DEVICE), target.to(DEVICE)
optimizer.zero_grad()
output = model(data)
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
model.eval()
correct = 0
with torch.no_grad():
for batch_idx, (data, target) in enumerate(valid_loader):
if batch_idx * BATCHSIZE >= N_VALID_EXAMPLES:
break
data, target = data.view(data.size(0), -1).to(DEVICE), target.to(DEVICE)
output = model(data)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).sum().item()
accuracy = correct / min(len(valid_loader.dataset), N_VALID_EXAMPLES)
trial.report(accuracy, epoch)
if trial.should_prune():
raise optuna.exceptions.TrialPruned()
return accuracy
if __name__ == "__main__":
study = optuna.create_study(
study_name="pytorch_example",
storage=os.environ["DATABASE_URL"],
load_if_exists=True,
direction="maximize",
)
study.optimize(objective, n_trials=100, timeout=600)tfkeras
While PyTorch is the go-to library for research, Keras with the TensorFlow backend is popular for its simplicity and ease of use. In this example, you will optimize the number of filters, kernel size, strides, activation functions, and learning rate in a convolutional neural network for the MNIST dataset.
import urllib
import os
import optuna
from tensorflow.keras.backend import clear_session
from tensorflow.keras.datasets import mnist
from tensorflow.keras.layers import Conv2D
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import Flatten
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import RMSprop
N_TRAIN_EXAMPLES = 3000
N_VALID_EXAMPLES = 1000
BATCHSIZE = 128
CLASSES = 10
EPOCHS = 10
def objective(trial):
clear_session()
(x_train, y_train), (x_valid, y_valid) = mnist.load_data()
img_x, img_y = x_train.shape[1], x_train.shape[2]
x_train = x_train.reshape(-1, img_x, img_y, 1)[:N_TRAIN_EXAMPLES].astype("float32") / 255
x_valid = x_valid.reshape(-1, img_x, img_y, 1)[:N_VALID_EXAMPLES].astype("float32") / 255
y_train = y_train[:N_TRAIN_EXAMPLES]
y_valid = y_valid[:N_VALID_EXAMPLES]
input_shape = (img_x, img_y, 1)
model = Sequential()
model.add(
Conv2D(
filters=trial.suggest_categorical("filters", [32, 64]),
kernel_size=trial.suggest_categorical("kernel_size", [3, 5]),
strides=trial.suggest_categorical("strides", [1, 2]),
activation=trial.suggest_categorical("activation", ["relu", "linear"]),
input_shape=input_shape,
)
)
model.add(Flatten())
model.add(Dense(CLASSES, activation="softmax"))
learning_rate = trial.suggest_float("learning_rate", 1e-5, 1e-1, log=True)
model.compile(
loss="sparse_categorical_crossentropy",
optimizer=RMSprop(learning_rate=learning_rate),
metrics=["accuracy"],
)
model.fit(
x_train,
y_train,
validation_data=(x_valid, y_valid),
shuffle=True,
batch_size=BATCHSIZE,
epochs=EPOCHS,
verbose=False,
)
score = model.evaluate(x_valid, y_valid, verbose=0)
return score[1]
if __name__ == "__main__":
study = optuna.create_study(
study_name="tfkeras_example",
storage=os.environ["DATABASE_URL"],
load_if_exists=True,
direction="maximize",
)
study.optimize(objective, n_trials=100, timeout=600)Creating the Docker Image
To run the hyperparameter tuning process in a Kubernetes cluster, you'll need to containerize your application using Docker by creating a Docker image. The Docker image will contain your Python code, dependencies, and the necessary configuration files to run.
FROM python:3.10-slim-buster
WORKDIR /usr/src/
RUN pip install --no-cache-dir optuna psycopg2-binary OTHER_DEPENDENCIES
COPY hyperparam_optimization.py .Depending on the machine learning library you're using, you'll need to install the appropriate dependencies in the Docker image. Each of the examples above requires the optuna, and psycopg2-binary packages, but you will need additional dependencies for each of the examples :
- For scikit-learn, you'll need to install
scikit-learn - For XGBoost, you'll need to install
xgboost - For PyTorch, you'll need to install
torchandtorchvision - For TensorFlow/Keras, you'll need to install
tensorflow
You'll want to build the Docker image later, once the Kubernetes cluster is set up, so that the image is available to the Kubernetes nodes.
Setting up Kubernetes
To run distributed hyperparameter tuning across multiple nodes, you'll need a Kubernetes cluster. For this guide, you'll use Minikube to set up a local Kubernetes cluster on your machine. Minikube is a lightweight Kubernetes distribution, making it easy to get started with Kubernetes development.
To start Minikube, run the following command:
minikube startThis command will create a new Kubernetes cluster using the default settings. Once Minikube is up and running, you can interact with the cluster using the kubectl command-line tool.
To check the status of your cluster, run:
kubectl cluster-infoThis command will display information about the Kubernetes cluster, including the API server address and the cluster services.
Now that minikube is running, you can build your Docker using:
eval "$(minikube docker-env)"
docker image build -t "optuna-kubernetes:example" .Note the eval "$(minikube docker-env)" command, which sets the Docker environment variables to point to the Minikube Docker daemon. This allows you to build the Docker image inside the Minikube cluster, making it available to the Kubernetes nodes.
To submit jobs to the Kubernetes cluster, you'll need to create a Kubernetes manifest file. This file describes the task you want to run, including the container image, command, and environment variables. You can define the number of parallel jobs to run and the restart policy for the job.
To allow the Job to access the Neon Postgres database, you'll need to create a Kubernetes Secret containing the database credentials. You can create the secret from a .env file containing the database URL like so:
kubectl create secret generic optuna-postgres-secrets --from-env-file=.envwhere your .env file contains the database URL from the Neon Console:
DATABASE_URL=YOUR_DATABASE_URLOr from the raw string in the Neon Console like so:
kubectl create secret generic optuna-postgres-secrets \
--from-literal=DATABASE_URL=YOUR_DATABASE_URLNow, you can create the Kubernetes Job manifest file:
---
apiVersion: batch/v1
kind: Job
metadata:
name: worker
spec:
parallelism: 3
template:
spec:
restartPolicy: OnFailure
containers:
- name: worker
image: optuna-kubernetes:example
imagePullPolicy: IfNotPresent
command:
- python
- hyperparam_optimization.py
envFrom:
- secretRef:
name: optuna-postgres-secretsHere, the manifest tells Kubernetes to launch 3 parallel jobs, each running the hyperparam_optimization.py script inside the Docker container, and if the job fails, Kubernetes will restart it automatically. The envFrom field specifies that the Job should use the optuna-postgres-secrets secret you just created to access the Neon Postgres database.
Finally, you can submit the Job to the Kubernetes cluster using the following command:
kubectl apply -f k8s-manifests.yamlNow, if you run the following command, you should see the Job running in the Kubernetes cluster:
kubectl get jobsand if you run the following command, you should see the 3 pods running the job:
kubectl get podsMonitoring
To monitor your Kubernetes cluster, you can use the Kubernetes Dashboard, a web-based UI for managing and monitoring your cluster. To access the Kubernetes Dashboard, run the following command:
minikube dashboardLikewise, you can monitor logs of the running pods to monitor using a tool like stern, which allows you to tail logs from multiple pods at once:
stern .Here, you can see that the first pod creates a new study, and the other pods join the existing study. Then, each pods runs its trial, logs the result, and creates new a trial based on the results of the previous ones in the database.
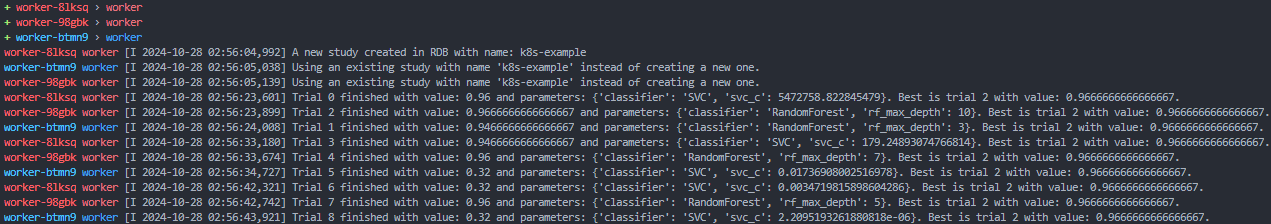
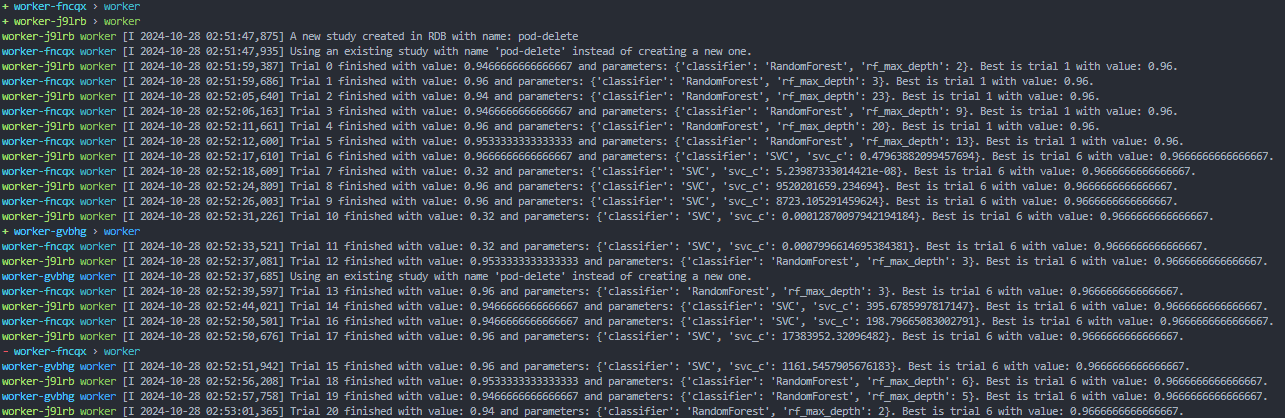
To show off the power of Kubernetes fault tolerance, you can delete one of the pods, and see that the job is automatically restarted on a new pod. First, find all the running pods and chose one to delete:
kubectl get podsThen, delete the pod:
kubectl delete pod <POD_NAME>In the stern logs, you can see the pod getting removed, and a new pod being created to replace it, all while continuing the same study as before.
Conclusion
Now, you have successfully set up distributed hyperparameter tuning using Optuna, Neon Postgres, and Kubernetes. By leveraging Kubernetes to manage multiple nodes running hyperparameter tuning jobs, you can speed up the optimization process and find the best hyperparameters for your machine learning models more efficiently. This kind of task, which sees bursts of database activity followed by long periods of inactivity, is well-suited to a serverless database like Neon Postgres, which can scale dynamically to any workload, then back to zero.
To take this to the next step, you can leverage cloud Kubernetes services like Azure Kubernetes Service (AKS) or Amazon Elastic Kubernetes Service (EKS). These services offer managed Kubernetes clusters that can scale to hundreds of nodes to run your jobs at scale. You can also integrate with cloud storage services like Azure Blob Storage or Amazon S3 to store your training data and model checkpoints, making it easier to manage large datasets and distributed training workflows.